US Pharm. 2022;47(4):4-12.
ABSTRACT: Opioids are among the most common medications used in the treatment of pain, and they carry many serious risks if used inappropriately. Pharmacogenomic considerations can impact a patient’s treatment plan for chronic pain, as CYP polymorphisms may interfere with nonsteroidal anti-inflammatory drugs, tricyclic antidepressants, and opioids. In particular, CYP2D6 polymorphisms may play a considerable part in the safety and efficacy of codeine, hydrocodone, and tramadol, as outlined in the Clinical Pharmacogenetic Implementation Consortium (CPIC) guideline on opioids. Based on current research, the CPIC recently updated its guidance on opioid use and interactions with CYP2D6, OPRM1, and COMT polymorphisms. Greater pharmacist understanding of CYP polymorphisms can lead to safer opioid prescribing and better outcomes for patients receiving pain management.
Pharmacogenomics is an emerging field of personalized medicine that tailors the use of drugs based on a patient’s genomic variations in order to maximize efficacy and minimize side effects.1 Currently, the most common pharmacogenomic testing is for detection of nucleotide variations in the genes encoding for CYP450 or phase II drug-metabolizing enzymes. Allelic variations in these enzymes lead to altered drug-metabolizing capacity in patients carrying the alleles. In pharmacogenomics, these variations in drug-metabolizing capacity are categorized as metabolic phenotypes.
Depending on whether a given enzyme metabolizes a drug to active or inactive metabolites, polymorphisms of that enzyme may lead to an increased risk of treatment failure or side effects.1 For example, if a drug is metabolized by an enzyme to inactive metabolites (e.g., celecoxib and CYP2C9), loss of function in the alleles of that enzyme will result in a poor (i.e., underactive) metabolizer phenotype, a higher concentration of the parent drug, and accordingly an increased risk of side effects. In comparison, an allele that causes increased metabolic activity of the same enzyme will result in a rapid or ultrarapid (overactive) metabolizer phenotype, a lower concentration of the drug, and the risk of therapeutic failure due to subtherapeutic plasma concentrations. The polymorphism has the opposite effect if the enzyme is responsible for activating a prodrug to its active metabolite for the therapeutic effect (e.g., codeine and CYP2D6).
Given the rapid decline in the cost of pharmacogenomic testing and growing clinical evidence on the impact of pharmacogenomics on pharmacotherapy, pharmacists are ideally positioned to identify how pharmacogenomics affects a given pharmacotherapy and to recommend and interpret pharmacogenomic test results in order to optimize therapy.
The Clinical Pharmacogenetics Implementation Consortium (CPIC) publishes guidelines on the impact of pharmacogenomics on a drug’s efficacy or risk of side effects based on prospective clinical evidence.2 Recommendations may include close monitoring, altering the dose, or avoiding a drug altogether. This article is a critical review of recently published CPIC guidelines on opioid use based on pharmacogenomic variations.3
Opioid Use in Pain Management
Opioids are among the most common medications used in the treatment of pain, and they carry many serious risks if used inappropriately. In the management of both acute and chronic noncancer pain, palliative care, or end-of-life pain, opioids are intended to be used after nonpharmacologic interventions and nonopioid pharmacologic treatment options have failed or proved insufficient.4 Nonpharmacologic treatment options that may be attempted prior to opioid therapy include cognitive-behavioral therapy, exercise therapy, physical therapy, and weight loss.4 For the treatment of active cancer pain, clinicians may initiate opioid therapy in addition to nonopioid analgesics when the pain is severe enough to warrant it.5 The use of opioids in oncology patients should be closely monitored for efficacy (in order to avoid lack of pain control) and safety.5 The many risks of opioid use, including sedation, constipation, dependence, overdose, and death, should be discussed with the patient.4 Opioid treatment should be continued only after the patient’s improvements in pain and function have been observed to outweigh these significant risks.4 In addition, the CDC recommends that clinicians consider developing an exit strategy if treatment with opioids does not prove beneficial for the patient.4
Meperidine is not recommended for the treatment of cancer pain because of the risk of toxicity, and given variations in metabolism, codeine is not preferred unless pharmacogenomic results are available.5 Pharmacologic nonopioid options include acetaminophen, nonsteroidal anti-inflammatory drugs (NSAIDs), antidepressants such as serotonin-norepinephrine reuptake inhibitors (SNRIs) and tricyclic antidepressants (TCAs), antiepileptic medications such as gabapentinoids, and topical analgesics such as lidocaine and capsaicin.
CYP2D6 Polymorphism
CYP2D6, which is one of the major CYP450 enzymes in human hepatocytes, is responsible for the metabolism of a variety of central nervous system (CNS)–active drugs, including many centrally acting analgesics such as opioids. The CYP2D6 gene is highly polymorphic, with single nucleotide allelic variations resulting in reduced or no enzyme activity. Additionally, CYP2D6 is unique in that it can have multiple copies of these alleles. The CPIC recommends a scoring system based on clinical studies to help translate CYP2D6 genotypes into phenotypes. In this scoring system, CYP2D6 alleles are given an activity score from 0 (no activity) to 1 (normal activity). The sum of the alleles’ activity scores constitutes the activity score, which corresponds to the patient’s ability to metabolize CYP2D6 substrates. In the presence of multiple copies, the allele activity score is multiplied by the number of CYP2D6 copies the patient has, and the sum of these scores is the patient’s activity score. Multiplication in genes with normal activity can result in higher activity scores and increased metabolism. Ultimately, CYP2D6 polymorphisms are categorized into “ultrarapid,” “normal,” “intermediate,” “poor,” and “indeterminate” metabolizer phenotypes.3,6
Impact of CYP2D6 Polymorphism on Opioid Metabolism
Several common opioids—specifically, codeine, hydrocodone, methadone, oxycodone, and tramadol—are metabolized by the CYP2D6 enzyme (FIGURE 1).7 CYP2D6 converts codeine and tramadol to their more active metabolites, morphine and O-desmethyltramadol, respectively.8 Therefore, alterations in the metabolism of these drugs can have a profound impact on their analgesic effect or risk of side effects. Oxycodone and hydrocodone are also converted by CYP2D6 into oxymorphone and hydromorphone, respectively.9 Although oxymorphone and hydromorphone have higher morphine equivalents than their parent drugs, both oxycodone and hydrocodone are active drugs, so alterations in their metabolism have a less profound clinical effect than codeine and tramadol. Methadone, although partially metabolized by CYP2D6, is predominantly metabolized by other enzymes.10
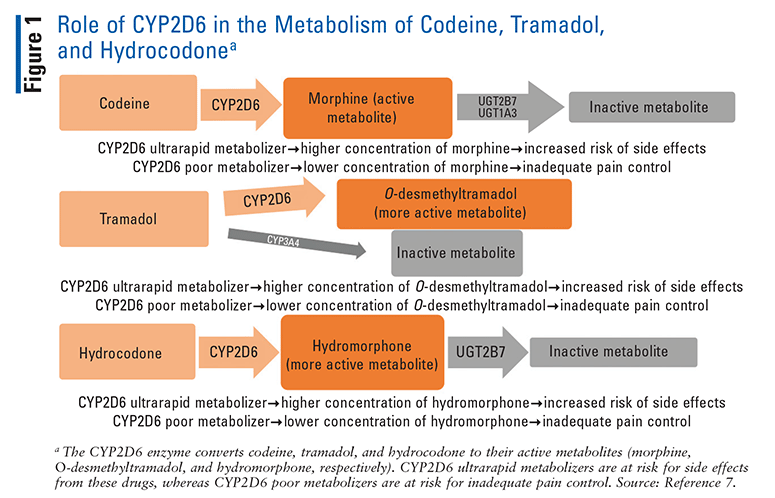
Evidence of CYP2D6 Polymorphisms’ Effect on Opioids and Clinical Recommendations
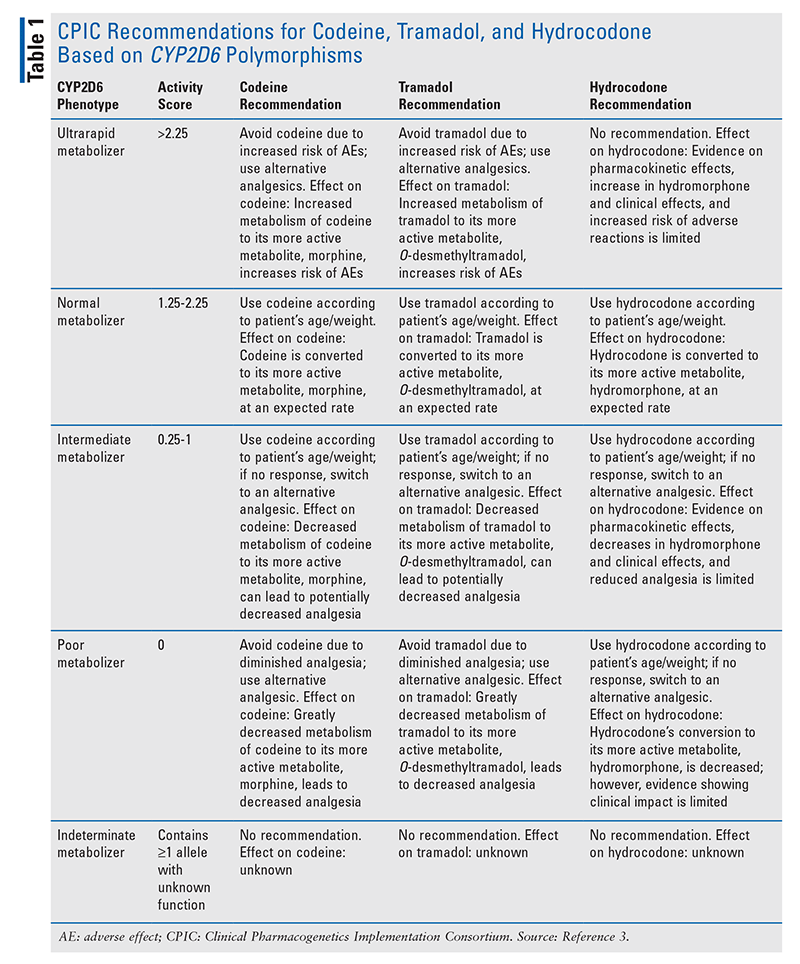
Among the opioids, the impact of CYP2D6 polymorphisms on codeine has been the best established. Poor metabolizers have shown both pharmacokinetic and clinical differences compared with normal metabolizers. One study showed that patients with poor-metabolizer genotypes who took codeine had 96% lower mean serum morphine AUC and 95% lower morphine serum peak concentrations compared with normal and intermediate metabolizers.11 This study also showed no analgesia in poor metabolizers compared with normal and intermediate metabolizers, who demonstrated analgesic response.12 Another study found that patients with sickle cell disease who did not respond to codeine therapy for pain management were more likely to have an activity score less than 1.5.13 The CPIC determined that the evidence was sufficient to conclude that CYP2D6 poor metabolizers who take codeine have greatly reduced morphine formation, thus resulting in decreased analgesia. Based on these clinical studies, the CPIC strongly recommended that codeine be avoided in CPY2D6 poor metabolizers.3
Alternatively, in pharmacokinetic studies, CYP2D6 ultrarapid metabolizers have been shown to have higher morphine levels.12 Ultrarapid metabolizers showed a 45% higher median plasma morphine AUC and 50% higher plasma concentrations of morphine compared with normal metabolizers.12 Clinical outcomes such as severe adverse effects (AEs) in ultrarapid metabolizers are limited to case reports.14 Based on these studies, the CPIC concluded definitively that when they take codeine, CYP2D6 ultrarapid metabolizers have increased morphine formation leading to increased AEs, and that they should avoid the use of codeine.3
Similarly, patients who were CYP2D6 poor metabolizers and took tramadol showed lower median plasma concentrations of O-desmethyltramadol compared with normal metabolizers.10 In several studies, CYP2D6 poor metabolizers had less analgesia after taking tramadol compared with normal metabolizers.10,15,16 These studies led the CPIC to definitively conclude that CYP2D6 poor metabolizers have decreased O-desmethyltramadol and less analgesia, and that they should avoid the use of tramadol.3
Ultrarapid metabolizers have demonstrated both higher O-desmethyltramadol AUC and greater analgesia compared with normal metabolizers.17 Additionally, case studies have reported the occurrence of severe AEs in ultrarapid metabolizers, leading the CPIC to definitively conclude that CYP2D6 metabolizers should avoid the use of tramadol because of the potential for toxicity caused by an increase in O-desmethyltramadol.3,18,19
Although hydromorphone plasma levels are similar between ultrarapid metabolizers and normal metabolizers who use hydrocodone, there is a discernible difference between poor metabolizers and normal metabolizers. Normal metabolizers administered hydrocodone had hydromorphone levels five times higher compared with poor metabolizers given the same dose.20 However, clinical studies highlighting the effects of CYP2D6 polymorphisms on hydrocodone safety and efficacy are limited. For this reason, the CPIC added an optional recommendation to switch analgesics only if a CYP2D6 poor metabolizer does not respond to label-based doses of hydrocodone.3
Recommendations for oxycodone and methadone in CYP2D6 gene polymorphisms are uncertain. Although in one study poor metabolizers taking oxycodone had oxymorphone levels that were 67% lower than in normal metabolizers, this did not necessitate an increase in oxycodone dose in poor metabolizers.21 Additionally, ultrarapid metabolizers showed no pharmacokinetic differences for oxymorphone levels compared with normal metabolizers.21,22 Prospective studies of clinical effectiveness are variable; some studies show analgesic and AE differences between poor, normal, and ultrarapid metabolizers with oxycodone use, whereas other studies demonstrate no difference, making evidence inconclusive.21-25 CYP2D6 metabolizes methadone to an inactive metabolite to a minor extent, but AEs and analgesia do not seem to be affected by CYP2D6 polymorphisms. For these reasons, the CPIC concluded that data are insufficient to make recommendations for oxycodone and methadone.
Current CPIC recommendations for opioid use based on CYP2D6 gene polymorphisms are summarized in TABLE 1.
Safer Alternatives and Other Considerations
For patients who are not candidates for codeine, tramadol, or hydrocodone based on their CYP2D6 polymorphisms, the CPIC recommends using opioids that are not metabolized by CYP2D6—namely, morphine, fentanyl, tapentadol, buprenorphine, and meperidine. When indicated, pharmacologic nonopioid options may include acetaminophen, NSAIDs, SNRIs, TCAs, gabapentinoids, and topical analgesics. When deciding on alternatives, clinicians should also consider other CYP interactions. For example, if genetic testing results are available for a patient, providers should be aware of alterations in the metabolism of CYP2C9 for NSAIDs and both CYP2D6 and CYP2C19 for TCAs.26
Pharmacogenomics-guided opioid therapy enables clinicians to determine the best options for avoiding treatment failure and serious side effects. However, clinicians should continue to personalize pain management by considering other factors as well (TABLE 2).2-4,26-34 For example, elderly patients are at higher risk for falls and fractures secondary to oversedation.4 Therefore, opioid therapy in elderly patients should be initiated and titrated with caution, and patients should be monitored more frequently for constipation, cognitive function, sedation, and fall risk.
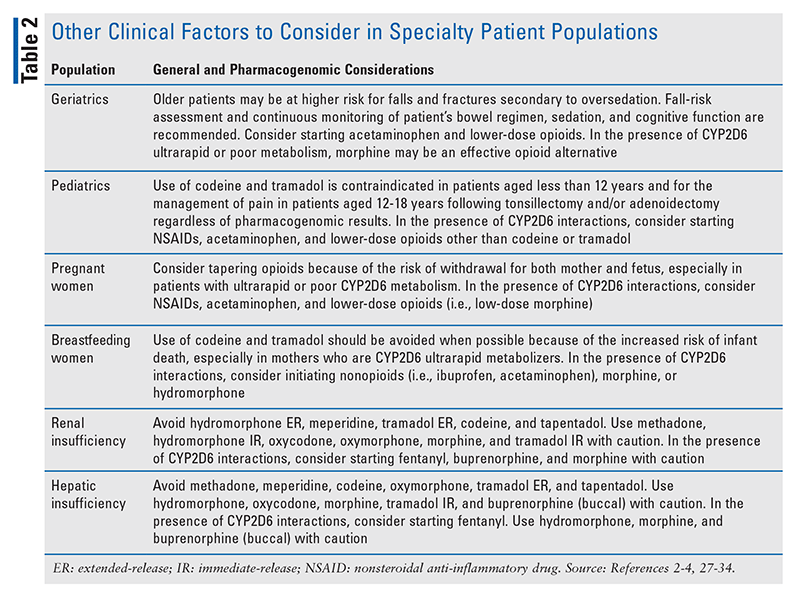
It should be noted that the risks of oversedation and respiratory depression are considerable if opioid therapy is used in conjunction with additional CNS depressants, such as muscle relaxants and benzodiazepines.4 Particular caution should be taken when CYP2D6 substrates are used in the presence of significant drug-drug interactions, especially CYP2D6 inhibitors (e.g., bupropion, cinacalcet, fluoxetine, paroxetine, quinidine) and inducers (e.g., dexamethasone, oritavancin, rifampin); this is because interacting medications can potentially change the CYP2D6 phenotype (i.e., phenoconversion).35 An alternative opioid independent of CYP2D6 may be safer in patients with interacting medications.
Impact of COMT and OPRM1 Polymorphism on Opioid Use
The recent CPIC opioid guideline introduces other genes that potentially may be associated with altered opioid efficacy and AEs—namely, OPRM1 (mu receptor) and catechol-O-methyltransferase (COMT).3 Because OPRM1 is a gene that codes for mu receptors, alterations in OPRM1 can cause reduced mu-receptor expression.36 COMT conjugates methyl to catecholamines, such as dopamine, and can affect synaptic levels of catecholamines and, in turn, pain perception. COMT polymorphism can lessen the enzyme’s ability to methylate dopamine, resulting in higher pain levels and decreased opioid effectiveness.37 Although polymorphisms in OPRM1 and COMT have been reported, study results on clinical differences in patients with OPRM1 and COMT polymorphism are inconsistent. Some studies show a slightly increased opioid requirement in patients with at least one copy of the rs1799971 G allele of OPRM1.3 No data demonstrate increased opioid AEs in patients with rs4680, a COMT variant. Therefore, insufficient evidence is currently available on the presence of OPRM1 and COMT allelic polymorphisms and opioid-dose requirements.3 Owing to the lack of strong evidence, the CPIC did not make any recommendations regarding opioid dosing based on OPRM1 or COMT genetic polymorphism. However, this likely will change as knowledge in this area expands.
The Pharmacist’s Role
Pharmacists play a critical role in the optimization of pain medications and risk mitigation in patients receiving chronic opioid therapy. Titrating nonopioid analgesics to their optimal tolerated doses can reduce the need to escalate opioid regimens or even to introduce them altogether. On a broader scale, pharmacists can play a vital part in curbing the national opioid crisis by providing patient and family education and either requesting a prescription for or furnishing naloxone to patients at higher risk. By better understanding the pathways of opioid metabolism, pharmacists can identify patients who would be appropriate candidates for CYP pharmacogenomic testing and can help select the optimal pharmacogenomic test that takes patients’ ethnicity and other clinical factors into consideration.38 For patients who undergo pharmacogenomic testing, pharmacists can take the lead in accurately interpreting the results and making recommendations for appropriate therapy. Using the test results, pharmacists may be able to identify patients with drug-seeking behavior related to uncontrolled pain and offer them alternatives. In patients without genetic testing who are responding unconventionally to opioid treatment, genetic alterations may help explain treatment failure and point to alternative options.
Conclusion
As medicine continues to become more personalized, prescribers must adjust to its advances. Opioids must be used safely and monitored closely. Pharmacogenomics helps practitioners predict clinical responses to therapies. Understanding the principles of genetic testing for opioids leads to therapy that yields improved responses and fewer AEs. Pharmacogenomic tests can help pharmacists identify patients at higher risk for opioid-related side effects or treatment failure.
REFERENCES
1. MedlinePlus. What is pharmacogenomics? https://medlineplus.gov/genetics/understanding/genomicresearch/pharmacogenomics/. Accessed December 22, 2021.
2. Clinical Pharmacogenetics Implementation Consortium. What is CPIC? https://cpicpgx.org/. Accessed December 22, 2021.
3. Clinical Pharmacogenetics Implementation Consortium. CPIC® guideline for opioids and CYP2D6, OPRM1, and COMT. https://cpicpgx.org/guidelines/guideline-for-codeine-and-cyp2d6/. Accessed December 22, 2021.
4. Dowell D, Haegerich TM, Chou R. CDC guideline for prescribing opioids for chronic pain—United States, 2016. MMWR Recomm Rep. 2016;65(No. RR-1):1-49.
5. Portenoy RK, Mehta Z, Ahmed E. Cancer pain management with opioids: optimizing analgesia. UpToDate. Waltham, MA: UpToDate Inc. www.uptodate.com. Accessed January 3, 2022.
6. Caudle KE, Sangkuhl K, Whirl-Carrillo M, et al. Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin Transl Sci. 2020;13(1):116-124.
7. Smith HS. Opioid metabolism. Mayo Clin Proc. 2009;84(7):613-624.
8. Stamer UM, Musshoff F, Kobilay M, et al. Concentrations of tramadol and O-desmethyltramadol enantiomers in different CYP2D6 genotypes. Clin Pharmacol Ther. 2007;82(1):41-47.
9. Lalovic B, Kharasch E, Hoffer C, et al. Pharmacokinetics and pharmacodynamics of oral oxycodone in healthy human subjects: role of circulating active metabolites. Clin Pharmacol Ther. 2006;79(5):461-479.
10. Kharasch ED. Current concepts in methadone metabolism and transport. Clin Pharmacol Drug Dev. 2017;6(2):125-134.
11. Eckhardt K, Li S, Ammon S, et al. Same incidence of adverse drug events after codeine administration irrespective of the genetically determined differences in morphine formation. Pain. 1998;76(1):27-33.
12. Brousseau DC, McCarver DG, Drendel AL, et al. The effect of CYP2D6 polymorphisms on the response to pain treatment for pediatric sickle cell pain crisis. J Pediatr. 2007;150(6):623-626.
13. Kirchheiner J, Schmidt H, Tzvetkov M, et al. Pharmacokinetics of codeine and its metabolite morphine in ultra-rapid metabolizers due to CYP2D6 duplication. Pharmacogenomics J. 2006;7(4):257-265.
14. Gasche Y, Daali Y, Fathi M, et al. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N Engl J Med. 2004;351(27):2827-2831.
15. Stamer UM, Lehnen K, Höthker F, et al. Impact of CYP2D6 genotype on postoperative tramadol analgesia. Pain. 2003;105(1-2):231-238.
16. Poulsen L, Arendt-Nielsen L, Brøsen K, Sindrup SH. The hypoalgesic effect of tramadol in relation to CYP2D6. Clin Pharmacol Ther. 1996;60(6):636-644.
17. Kirchheiner J, Keulen JT, Bauer S, et al. Effects of the CYP2D6 gene duplication on the pharmacokinetics and pharmacodynamics of tramadol. J Clin Psychopharmacol. 2008;28(1):78-83.
18. Elkalioubie A, Allorge D, Robriquet L, et al. Near-fatal tramadol cardiotoxicity in a CYP2D6 ultrarapid metabolizer. Eur J Clin Pharmacol. 2011;67(8):855-858.
19. Stamer UM, Stüber F, Muders T, Musshoff F. Respiratory depression with tramadol in a patient with renal impairment and CYP2D6 gene duplication. Anesth Analg. 2008;107(3):926-929.
20. Otton SV, Schadel M, Cheung SW, et al. CYP2D6 phenotype determines the metabolic conversion of hydrocodone to hydromorphone. Clin Pharmacol Ther. 1993;54(5):463-472.
21. Zwisler ST, Enggaard TP, Mikkelsen S, et al. Impact of the CYP2D6 genotype on post-operative intravenous oxycodone analgesia. Acta Anaesthesiol Scand. 2010;54(2):232-240.
22. Andreassen TN, Eftedal I, Klepstad P, et al. Do CYP2D6 genotypes reflect oxycodone requirements for cancer patients treated for cancer pain? A cross-sectional multicentre study. Eur J Clin Pharmacol. 2012;68(1):55-64.
23. Zwisler ST, Enggaard TP, Noehr-Jensen L, et al. The hypoalgesic effect of oxycodone in human experimental pain models in relation to the CYP2D6 oxidation polymorphism. Basic Clin Pharmacol Toxicol. 2009;104(4):335-344.
24. Samer CF, Daali Y, Wagner M, et al. Genetic polymorphisms and drug interactions modulating CYP2D6 and CYP3A activities have a major effect on oxycodone analgesic efficacy and safety. Br J Pharmacol. 2010;160(4):919-930.
25. Klimas R, Witticke D, El Fallah S, Mikus G. Contribution of oxycodone and its metabolites to the overall analgesic effect after oxycodone administration. Expert Opin Drug Metab Toxicol. 2013;9(5):517-528.
26. Kathuria A, Roosan MR, Sharma A. CYP2C9 polymorphism and use of oral nonsteroidal anti-inflammatory drugs. US Pharm. 2021;54(3):23-30.
27. Nucynta ER (tapentadol) package insert. Stoughton, MA: Collegium Pharmaceutical, Inc; March 2021.
28. Nucynta (tapentadol) package insert. Stoughton, MA: Collegium Pharmaceutical, Inc; March 2021.
29. Belbuca (buprenorphine buccal film) package insert. Raleigh, NC: BioDelivery Sciences International, Inc; March 2021.
30. American College of Obstetricians and Gynecologists’ Committee on Clinical Consensus–Obstetrics. Pharmacologic stepwise multimodal approach for postpartum pain management: ACOG Clinical Consensus no. 1. Obstet Gynecol. 2021;138(3):507-517.
31. Morphine. Drugs and lactation database (LactMed) [Internet]. Bethesda, MD: National Library of Medicine; 2022. www.ncbi.nlm.nih.gov/books/NBK501237/. Accessed February 11, 2022.
32. Morphine: drug information. UpToDate. Waltham, MA: UpToDate Inc. www.uptodate.com. Accessed January 3, 2022.
33. Opana (oxymorphone) package insert. Malvern, PA: Endo Pharmaceuticals, Inc; October 2019.
34. Opana ER (oxymorphone) package insert. Malvern, PA: Endo Pharmaceuticals, Inc; October 2019.
35. Flockhart DA, Thacker D, McDonald C, Desta Z. The Flockhart cytochrome P450 drug-drug interaction table. Division of Clinical Pharmacology, Indiana University School of Medicine (updated 2021). https://drug-interactions.medicine.iu.edu/. Accessed February 11, 2022.
36. Zhang Y, Wang D, Johnson AD, et al. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem. 2005;280(38):32618-32624.
37. Zubieta JK, Heitzeg MM, Smith YR, et al. COMT val158met genotype affects mu-opioid neurotransmitter responses to a pain stressor. Science. 2003;299(5610):1240-1243.
38. Sayer M, Duche A, Nguyen TJ, et al. Clinical implications of combinatorial pharmacogenomic tests based on cytochrome P450 variant selection. Front Genet. 2021;12:719671.
The content contained in this article is for informational purposes only. The content is not intended to be a substitute for professional advice. Reliance on any information provided in this article is solely at your own risk.
To comment on this article, contact rdavidson@uspharmacist.com.





